Table of Contents
Xmaris
Xmaris is a small computational cluster at the Lorentz Institute financed by external research grants. As such, its access is granted primarily to the research groups who have been awarded the grants. Other research groups wishing to use xmaris can enquire whether there is any left-over computing time by getting in touch with either
| Xavier Bonet Monroig | Oort 260 |
| Carlo Beenakker | Oort 261 |
to discuss what resources can be made available to their needs. After a preliminary assessment and approval, access to xmaris will be granted by the IT staff. Any technical questions should be addressed via https://helpdesk.lorentz.leidenuniv.nl to
| Leonardo Lenoci | HL409b |

Xmaris is optimised for multithreading applications and embarrassingly parallel problems, but there have been some recent investments to improve nodes interconnection communications to enable multiprocessing. Currently, multiprocessing is possible on the nodes of the ibIntel partition which are interconnected via an InfiniBand EDR switch. Each one of these nodes is capable of a practical 9.6 TFLOPS.
Xmaris is the successor of the maris cluster, renamed with a prefix x because its nodes deployment is automated using the xCAT software. Less formally, the presence of the x prefix also suggests the time of the year when xmaris was first made available to IL users, that is Christmas (Xmas).
Xmaris features and expected cluster lifetime
Xmaris runs CentOS v7 and consists for historical reasons of heterogeneous computation nodes. A list of configured nodes and partitions on the cluster can be obtained on the command line using slurm's sinfo.

Features that also describe the type of CPUs mounted in that node. To request allocation of specific features to the resource manager, see this example.
You can display just one node specs and features or all nodes specs and features with sinfo
# specific node sinfo -o " %n %P %t %C %z %m %f" -N -n maris077 # all nodes sinfo -o " %n %P %t %C %z %m %f" -N # all nodes (more concise) sinfo -Nel
Xmaris aims to offer a stable computational environment to its users in the period Dec 2019 – Jan 2024. Within this period, the OS might be patched only with important security updates. Past January 2024, all working xmaris nodes will be re-provisioned from scratch with newer versions of the operating system and software infrastructure. At this time all data stored in the temporary scratch disks will be destroyed and the disks reformatted.
Compute nodes data disks
All compute nodes have at least access to the following data partitions
| Mount Point | Type | Notes |
|---|---|---|
| /scratch | HD | temporary, local |
| /marisdata | NetApp | 2TB/user quota, medium-term storage, remote |
| /home | NetApp | 10GB/user quota, medium-term storage, remote |
| /ilZone/home | iRODS | 20GB/user quota, archive storage, remote |
Extra efficient scratch spaces are available to all nodes on the infiniband network (ibIntel)
| Mount Point | Type | Notes |
|---|---|---|
| /IBSSD | SSD | DISCONTINUED, InfiniBand/iSER1) |
| /PIBSSD | SSD | temporary, InfiniBand/BeeGFS |
Backup snapshots of /home are taken hourly, daily, and weekly and stored in /home/.snapshot/.
xmaris users are strongly advised they delete (or at least move to the shared data disk), if any, their data from the compute nodes scratch disks upon completion of their calculations. All data on the scratch disks might be cancelled without prior notice.
Note that disk policies might change at any time at the discretion of the cluster owners.
Please also note the following
- xmaris' home disk is different than your IL workstation or remote workspace home disk.
- The OLD (as in the old maris)
/clusterdatais deliberately made unavailable on xmaris, because it is no longer maintained. If you have any data on it, it is your responsibility to create backups. All data on/clusterdatawill get permanently lost in case of hardware failure.
Xmaris usage policies
Usage policies are updated regularly in accordance with the needs of the cluster owners and may change at any time without notice. At the moment there is an enforced usage limit of 128 CPUs per user that does not apply to the owners. Job execution priorities are defined via a complex multi-factor algorithm whose parameters can be displayed on the command line via
scontrol show config | grep -i priority
Xmaris live state
To monitor live usage of Xmaris you can either
- execute slurm's
sinfo(this requires shell access to the cluster, see below) - browse to https://xmaris.lorentz.leidenuniv.nl/ganglia/ from any IL workstation
How to access Xmaris
Access to Xmaris is not granted automatically to all Lorentz Institute members. Instead, a preliminary approval must be granted to you by the cluster owners (read here).
Once you have been authorised to use Xmaris, there are two ways to access its services:
- using a web browser (Strongly advised to the novel HPC user)
- using an SSH client (Expert users)
Both methods can provide terminal access, but connections via web browsers offer you extra services such as sftp (drag-and-drop file transfers), jupyter interactive notebooks, virtual desktops and more at the click of your mouse. We advise all users either unfamilair with the GNU/Linux terminal or new to HPC to use a web browser to interact with Xmaris.
Access via an ssh client
The procedure differs on whether you try to connect with a client connected to the IL intranet or not.
- When within the IL network, for instance if you are using a Lorentz Institute workstation, you have direct access to Xmaris. Open a terminal and type the command below (substitute username with your own IL username)
ssh xmaris.lorentz.leidenuniv.nl -l username
- When outside the IL network, for instance from home or using a wireless connection, you must first initiate an ssh tunnel to our SSH server and then connect to Xmaris
ssh -o ProxyCommand="ssh -W %h:%p username@styx.lorentz.leidenuniv.nl" username@xmaris.lorentz.leidenuniv.nl
 If you were a maris user prior to the configuration switch to xmaris, you might find out that many terminal functions and programs could not be working as expected. This is due to the presence in your xmaris home directory of old shell initialisation scripts still tied to the STRW sfinx environment. You can override them (preferably after making a backup copy) by replacing their contents with the default CentOS shell initialisation scripts, for instance for bash these are located in If you were a maris user prior to the configuration switch to xmaris, you might find out that many terminal functions and programs could not be working as expected. This is due to the presence in your xmaris home directory of old shell initialisation scripts still tied to the STRW sfinx environment. You can override them (preferably after making a backup copy) by replacing their contents with the default CentOS shell initialisation scripts, for instance for bash these are located in /etc/skel/.bashrc and /etc/skel/.bash_profile |
Web access
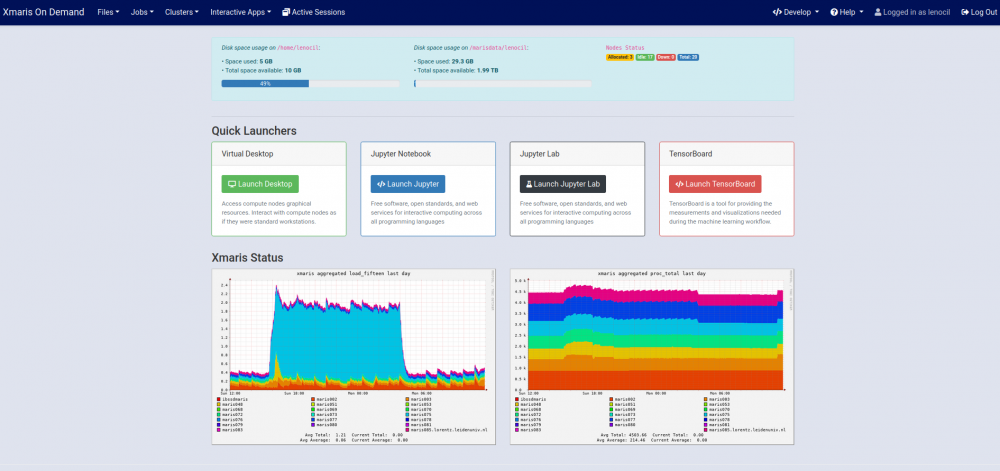
Xmaris services, that is terminal, scheduler/resource manager, jupyter notebooks and monitoring facilities, can be accessed easily via a browser without the need of additional plugins navigating to xmaris OpenOnDemand.
Similarly to a traditional shell access, Xmaris OpenOnDemand is available only for connections within the IL intranet. IL users who wish to access OpenOnDemand from their remote home locations could for example use the IL VPN or instruct their browsers to SOCKS-proxy their connections via our SSH server. Open a local terminal and type (substitute username with your IL username)
ssh -ND 7777 username@ssh.lorentz.leidenuniv.nl
then in your browser settings find the tab relative to the connection type and instruct the browser to use the SOCKS proxy located at localhost:7777 to connect to the internet. Alternatively, use the Lorentz Institute Remote Workspace.
Xmaris OnDemand allows you to
- Create/edit files and directories.
- Submit batch jobs to the slurm scheduler/resource manager.
- Open a terminal.
- Launch interactive applications such as jupyter notebooks, tensorboard, virtual desktops, etc..
- Monitor cluster usage.
- Create and launch your very own OnDemand application (read here).

Xmaris Partitions
| Partition | Number nodes | Timelimit | Notes |
|---|---|---|---|
| compAMD* | 6 | 15 days | |
| compAMDlong | 3 | 60 days | |
| compIntel | 2 | 5 days and 12 hours | |
| gpuIntel | 1 | 3 days and 12 hours | GPU |
| ibIntel | 8 | 7 days | InfiniBand, Multiprocessing |
*: default partition
Xmaris GPUs
| Node | Partition | GPUs | CUDA compatibility |
|---|---|---|---|
| maris075 | gpuIntel | 2 x Nvidia Tesla P100 16GB | 6.0 |
Xmaris GPUs must be allocated using slurm's –gres option, for instance
srun -p gpuIntel --gres=gpu:1 --pty bash -i
Xmaris scientific software
Xmaris uses EasyBuild to provide a build environment for its (scientific) software. Pre-installed software can be explored by means of the module spider command. For instance, you can query the system for all modules whose name starts with `mpi' by executing module -r spider '^mpi'. Installed softwares include
| GCC | GNU Compiler Collection |
| OpenBLAS | Basic Linear Algebra Subprograms |
| LAPACK | Linear Algebra PACKage |
| ScaLAPACK | Scalable Linear Algebra PACKage |
| CUDA | Compute Unified Device Architecture |
| FFTW | Fastest Fourier Transform in the West |
| EasyBuild | Software Build and Installation Framework |
| GSL | GNU Scientific Library |
| HDF5 | Management of Extremely Large and Complex Data Collections |
| git | Distributed Version Control System |
| Java | General-Purpose Programming Language |
| Miniconda | Free Minimal Installer for Conda |
| OpenMPI | Open Source Message Passing Interface Implementation |
| Python | Programming Language |
| PyCUDA | Python wrapper to CUDA |
| Perl | Programming Language |
| R | R is a Language and Environment for Statistical Computing and Graphics |
| Singularity | Containers software |
| Tensorflow | Machine Learning Platform |
| plc | The Planck Likelihood Code |
| cobaya | A code for Bayesian analysis in Cosmology |
| Clang | C language family frontend for LLVM |
| Graphviz | Graph visualization software |
| Octave | GNU Programming language for scientific computing |
| Mathematica* | Technical computing system |
* Usage of proprietary software is discouraged.
For an up-to-date list of installed software use the module avail command.
Any pre-installed software can be made available in your environment via the module load <module_name> command.
It is possible to save a list of modules you use often in a module collection to load them just with one command
module load mod1 mod2 mod3 mod4 mod5 mod6
modules save collection1
module restore collection1
# list collections
module savelist
TensorFlow Notes
Xmaris has multiple modules that provide TensorFlow. See ml avail TensorFlow.
| Module | Hardware | Partition | Additional Ops |
|---|---|---|---|
| TensorFlow/2.1.0-fosscuda-2019b-Python-3.7.4 | CPU, GPU | gpuIntel | TensorFlow Quantum |
| TensorFlow/1.12.0-fosscuda-2018b-Python-3.6.6 | CPU, GPU | gpuIntel | |
| TensorFlow-1.15.0-Miniconda3/4.7.10 | CPU | All |
The following example shows how you can create a tensorflow-aware jupyter notebook kernel that you can use for instance via the OpenOnDemand interface
# We use maris075 (GPU node) and load the optimised tf module ml load TensorFlow/2.1.0-fosscuda-2019b-Python-3.7.4 # We install ipykernel, because necessary to run py notebooks python -m pip install ipykernel --user # We create a kernel called TFQuantum based on python from TensorFlow/2.1.0-fosscuda-2019b-Python-3.7.4 python -m ipykernel install --name TFQuantum --display-name "TFQuantum" --user # We edit the kernel such that it does not execute python directly # but via a custom wrapper script cat $HOME/.local/share/jupyter/kernels/tfquantum/kernel.json { "argv": [ "/home/lenocil/.local/share/jupyter/kernels/tfquantum/wrapper.sh", "-m", "ipykernel_launcher", "-f", "{connection_file}" ], "display_name": "TFQuantum", "language": "python", "metadata": { "debugger": true } } # The wrapper script will call python but only after loading any # appropriate module cat /home/lenocil/.local/share/jupyter/kernels/tfquantum/wrapper.sh #!/bin/env bash ml load TensorFlow/2.1.0-fosscuda-2019b-Python-3.7.4 exec python $@ # DONE. tfquantum will appear in the dropdown list of kernels # upon creating a new notebook
TensorFlow with Graphviz
ml load TensorFlow/2.1.0-fosscuda-2019b-Python-3.7.4 pip install --user pydot ml load Graphviz/2.42.2-foss-2019b-Python-3.7.4 python -c "import tensorflow as tf;m = tf.keras.Model(inputs=[], outputs=[]);tf.keras.utils.plot_model(m, show_shapes=True)"
Installing extra software
If you need to run a software that is not present on Xmaris, you might:
- Request its installation via https://helpdesk.lorentz.leidenuniv.nl
- Install it yourself
- via EasyBuild (see instructions below on how to setup your personal EasyBuild environment)
- via a traditional configure/make procedure
Whatever installation method you might choose, please note that you do not have administrative rights to the cluster.
Installing software via EasyBuild

In order to use EasyBuild to build a software, you must first set up your development environment. This is usually done by
- Loading the EasyBuild module
- Indicating a directory in which to store your EasyBuild-built softwares
- Specifying EasyBuild's behaviour via EASYBUILD_* environment variables
- Build a software
In their simplest form, the steps outlined above can be translated into the following shell commands
module load EasyBuild mkdir /marisdata/<uname>/easybuild export EASYBUILD_PREFIX=/marisdata/<uname>/easybuild export EASYBUILD_OPTARCH=GENERIC eb -S ^Miniconda eb Miniconda2-4.3.21.eb -r
 The environment variable The environment variable EASYBUILD_OPTARCH instructs EasyBuild to compile software in a generic way so that it can be used on different CPUs. This is rather convenient in heterogeneous clusters such as xmaris to avoid recompilations of the same softwares on different compute nodes. This convenience comes of course at a cost; the executables so produced will not be as efficient as they would be on a given CPU. For more info read here. |
 When compiling OpenBLAS it is not sufficient to define When compiling OpenBLAS it is not sufficient to define EASYBUILD_OPTARCH to GENERIC to achieve portability of the executables. Some extra steps must be taken as described in https://github.com/easybuilders/easybuild/blob/master/docs/Controlling_compiler_optimization_flags.rst. A list of targets supported by OpenBLAS can be found here. |
Then execute
module use /marisdata/<uname>/easybuild/modules/all
to make available to the module comamnd any of the softwares built in your EasyBuild userspace.
 module use <path> will prepend <path> to your MODULEPATH. Should you want to append it instead, then add the option -a. To remove <path> from MODULEPATH execute module unuse <path>. |
Should you want to customise the building process of a given software please read how to implement EasyBlocks and write EasyConfig files or contact Leonardo Lenoci (HL409b).
Working with conda modules
Several conda modules are ready-to-use on maris. A possible use of these could be to clone and extend them with your packages of choice. Mind though that if you run conda init, conda will modify your shell initialisation scripts (e.g. ~/.bashrc) to load automatically the chosen conda environment.
This causes several problems in all cases in which you are supposed to work in a clean environment.
The steps below show as an example how you could skip conda init when activating a conda environment.
> ml load Miniconda3/4.7.10
> # note that if you specify prefix, you cannot specify the name
> conda create [--prefix <location where there is plenty of space and you can write to>] [--name TEST]
# the following fails
> conda activate TEST
CommandNotFoundError: Your shell has not been properly configured to use 'conda activate'.
To initialize your shell, run
$ conda init <SHELL_NAME>
Currently supported shells are:
- bash
- fish
- tcsh
- xonsh
- zsh
- powershell
See 'conda init --help' for more information and options.
IMPORTANT: You may need to close and restart your shell after running 'conda init'.
## do this instead
> source activate TEST
> # or if you used the --prefix option to crete the env
> # source activate <location where there is plenty of space and you can write to>
> ...
> conda deactivate
How to run a computation on Xmaris
Xmaris runs the slurm scheduler and resource manager. Computation jobs must be submitted as batch jobs or be run interactively via slurm. Any other jobs will be terminated without prior notice. Because this is not a slurm manual, you are encouraged to learn the basics by reading the slurm manual. Here we only give you a few simple examples.
Batch jobs
Batch jobs are computation jobs that do not execute interactively.
To submit a batch job to Xmaris' slurm you must first create a shell script which contains enough instructions to request the needed resources and to execute your program. The script can be written in any known interpreter to the system. Slurm instructions are prefixed by the chosen interpreter comment symbol and the word SBATCH.
An example bash-batch script that will request Xmaris to execute the program hostname on one node is
cat test.sh #!/bin/env bash #SBATCH --job-name=super #SBATCH --ntasks=1 #SBATCH --mem=1000 srun hostname
Batch scripts are then submitted for execution via sbatch
sbatch test.sh sbatch: Submitted batch job 738279474774293
and their status [PENDING|RUNNING|FAILED|COMPLETED] checked using squeue. You can recur to the command sstat to display useful information about your running job, such as memory consumption etc…
ssh shell access to an executing node is automatically granted by slurm and can also be used for debugging purposes.
Please consult the slurm manual for all possible sbatch options.
Interactive jobs
Interactive jobs are usually used for debugging purposes and in those cases in which the computation requires human interaction. Using interactive sessions you can gain shell access to a computation node
srun --pty bash -i
or execute an interactive program
srun -p compIntel -N1 -n 1 --mem=4000 --pty python -c "import sys; data = sys.stdin.readlines(); print(data)" -i Hello world ^D ['Hello world\n']
Parallelism 101
A parallel job runs a calculation whose computational subtasks are run simultaneously. The underlying principle is that
large computations could be more efficient if divided into smaller ones. Note however, that parallelism can in fact decrease the efficiency of a poorly written code in which communication and synchronisation between the different subtasks are not handled properly.
Parallelism is usually achieved either by
- multithreading (shared memory) on multiple cores of a single node
- multiprocessing (distributed memory) on multiple nodes
Multithreading
In multithreading programming all computation subtasks (threads) exist within the context of single process and share the process' resources. Threads are able to execute independently and are assigned by the operating system to multiple CPU cores and/or multiple CPUs effectively speeding up your calculation.
Multithreading can be achieved using libraries such as pthread and OpenMP.
Multiprocessing
Multiprocessing usually refers to computations subdivided into tasks that run on multiples nodes. This type of programming increases the resources available (e.g. more memory) to your computation by employing several nodes at the same time. MPI (Message Parsing Interface) defines the standards (in terms of syntax and rules) to implement multiprocessing in your codes.
MPI-enabled applications spawn multiple copies of the program, also called ranks, mapping each one of them to a processor. A computation node has usually multiple processors. The MPI interface lets you manage the allocated resources and the communication and synchronisation of the ranks.
It is easy to imagine how inefficient can be a poorly-written MPI application or an MPI application running on a cluster with slow nodes interconnects.
Hybrid Applications
This term refers to applications that use simultaneously multiprocessing (MPI) and multithreading (OpenMP).
How to launch a jupyter notebook
To launch a jupyter notebook login to xmaris OnDemand, select Interactive Apps –> Jupyter Notebook and specify the resources needed in the form provided, push Launch and wait until the notebook has launched.
Now you can interact with your notebook (click on Connect to Jupyter), open a shell on the executing node (click on Host >_hostaname), and analyse notebook log files for debugging purposes (click on Session ID xxxx-xxxx-xxxxx-xxxxxxx-xxx-xx).
If your notebook does not launch in a few seconds take the following actions
- Check the status of your jobs in the queue with
squeu -u <username>. - Examine the notebook log files (click on
Session ID xxxx-xxxx-xxxxx-xxxxxxx-xxx-xx). - If the suggestions above do not help, contact
support.
How to launch a jupyter notebook that uses GPUs
Repeat the steps above but make sure you select an appropriate GPU partition. Moreover, you must add an appropriate CUDA module to the field Extra modules needed otherwise the connection to the GPUs might not work as expected. For a list of cuda modules you could type in a terminal ml spider CUDA.
The form field Extra modules needed can accept more than one module as long as the modules names are separated by a space.

export NOTEBOOKDIR=/marisdata/$LOGNAME in your .bashrc

Launching jupyterlab instead of jupyter notebook
If you prefer the newest jupyter notebook features and interface, that is jupyetrlab, just proceed as above and after clicking on Connect to Jupyter replace the string tree with the string lab in the URL bar of your browser. For instance, a typical jupyterlab interface URL could look like this
https://xmaris.lorentz.leidenuniv.nl:4433/node/maris051.lorentz.leidenuniv.nl/26051/lab?
Custom jupyter kernels
A jupyter notebook kernel defines the notebook interpreter, such as python, R, matlab, etc…
It is possible to define custom kernels, for instance for a particular version of python including additional packages. The example below show how to install a python v3 kernel containing some additional python packages that you will then be able to use in your notebooks.
Create a python v3.5 kernel with additional numpy pkg
You can list all already available kernels
jupyter kernelspec list
then proceed to create a new one, for instance
module load Miniconda3/4.7.10 conda create --name py35 python=3.5 # default location in $HOME/.conda/envs/py35 source activate py35 conda install ipykernel ipython kernel install --name=py35kernel --user Installed kernelspec py35kernel in $HOME/.local/share/jupyter/kernels/py35kernel conda install h5py source deactivate py35

conda is a full package manager and environment management system and as such it might perform poorly in large environments.
Launch a jupyter notebook as described above and select the newly created py35kernel as shown in the figure below
numpy will also be available

# first remove the kernel source activate py35 jupyter kernelspec list jupyter kernelspec uninstall py35 source deactivate py35 # then delete the kernel environment conda env remove --name py35
Install a mathematica (wolfram) kernel
You can set it up following these notes https://github.com/WolframResearch/WolframLanguageForJupyter or follow these steps for a preconfigured setup.
- Open an SSH connection to xmaris
- Run
/marisdata/WOLFRAM/WolframLanguageForJupyter/configure-jupyter.wls add - The
wolframlanguageis now available among your kernels
Debugging jupyter lab/notebook sessions
xmaris OpenOnDemand writes jupyter sessions logs to subdirectories located in $HOME/ondemand/data/sys/dashboard/batch_connect/sys/jupyter/output/. Before contacting the helpdesk you are advised you analyse the contents of these subdirectories (one for each session) in particular the files called output.log and config.py.

xmaris slurm tips

xmaris runs the scheduler and resource manager slurm v18.08.6-2. Please consult the official manual for detailed information.

Here we report a few useful commands and their outputs to get you started. For the inpatients, look at the following slurm batch-script generator (NO RESPONSIBILITIES assumed! Study the script before submitting it.) which is available only from withing UL IPs.
Determine your slurm account name
sacctmgr show users <username>
Display detailed information about a node
sinfo -o " %n %P %t %C %z %m %f %l %L" -N -n maris077
Display detailed information about all nodes
sinfo -o " %n %P %t %C %z %m %f %G %l %L" -N
Launch an interactive session on a node
srun -w maris047 --pty bash -i
Display status of your jobs
squeue -u <username>
Interactive use of GPUs
srun -p gpuIntel --gres=gpu:1 --pty bash -i
Request nodes with particular features
srun --constraint="opteron&highmem" --pty bash -i
Run a multiprocessing application
Xmaris supports OpenMPI in combination with slurm's srun and infiniband on all nodes in the ibIntel partition.
First of all, make sure that the max locked memory is set to unlimited for your account by executing
# ulimit -l
unlimited
If that is NOT the case, please contact the IT support.
To run an MPI application see the session below
# login to the headnode and request resources $ salloc -N6 -n6 -p ibIntel --mem 2000 salloc: Granted job allocation 564086 salloc: Waiting for resource configuration salloc: Nodes maris[078-083] are ready for job # load the needed modules for the app to run $ ml load OpenMPI/4.1.1-GCC-10.3.0 OpenBLAS/0.3.17-GCC-10.3.0 # execute the app (note that the default MPI is set to pmi2) $ srun ./mpi_example Hello world! I am process number: 5 on host maris083.lorentz.leidenuniv.nl 11.000000 -9.000000 5.000000 -9.000000 21.000000 -1.000000 5.000000 -1.000000 3.000000 Hello world! I am process number: 4 on host maris082.lorentz.leidenuniv.nl 11.000000 -9.000000 5.000000 -9.000000 21.000000 -1.000000 5.000000 -1.000000 3.000000 Hello world! I am process number: 2 on host maris080.lorentz.leidenuniv.nl 11.000000 -9.000000 5.000000 -9.000000 21.000000 -1.000000 5.000000 -1.000000 3.000000 Hello world! I am process number: 1 on host maris079.lorentz.leidenuniv.nl 11.000000 -9.000000 5.000000 -9.000000 21.000000 -1.000000 5.000000 -1.000000 3.000000 Hello world! I am process number: 3 on host maris081.lorentz.leidenuniv.nl 11.000000 -9.000000 5.000000 -9.000000 21.000000 -1.000000 5.000000 -1.000000 3.000000 Hello world! I am process number: 0 on host maris078.lorentz.leidenuniv.nl 11.000000 -9.000000 5.000000 -9.000000 21.000000 -1.000000 5.000000 -1.000000 3.000000
Suggested readings
Requesting help
Please use this helpdesk form or email support.